Surrogate models for discrete predictions
Andre Weiner
TU Dresden, Institute of fluid mechanics, PSM

These slides and most
of the linked resources are licensed under a
Creative Commons Attribution 4.0
International License.
Last lecture
Deep learning
- test case overview
- simple feed-forward networks
- preparing the datasets
- generic training loop
- more building blocks
- dealing with uncertainty
- visualizing prediction errors
Outline
Predicting the stability regime of rising bubbles
- Scale-up approach
- Forces acting on rising bubbles
- Loading, inspecting, and preparing the data
- Binary classification
- Multi-class classification
Scale-up approach
Gas-liquid reactors
micro reactor
size: millimeter
source: SPP 1740
prediction of
- mass transfer
- enhancement
- mixing
- conversion
- selectivity
- yield
- ...

bubble column reactor
size: meter
source: R. M.
Raimundo, ENI
Scale-up modeling approach for bubble columns:
- below continuum scale ...
- boundary layers at bubbles ($\mu m$)
- resolved individual bubbles ($mm$)
- bubbles as point particles ($cm$)
- gas-liquid mixture ($m$)
How to transfer information between scales?
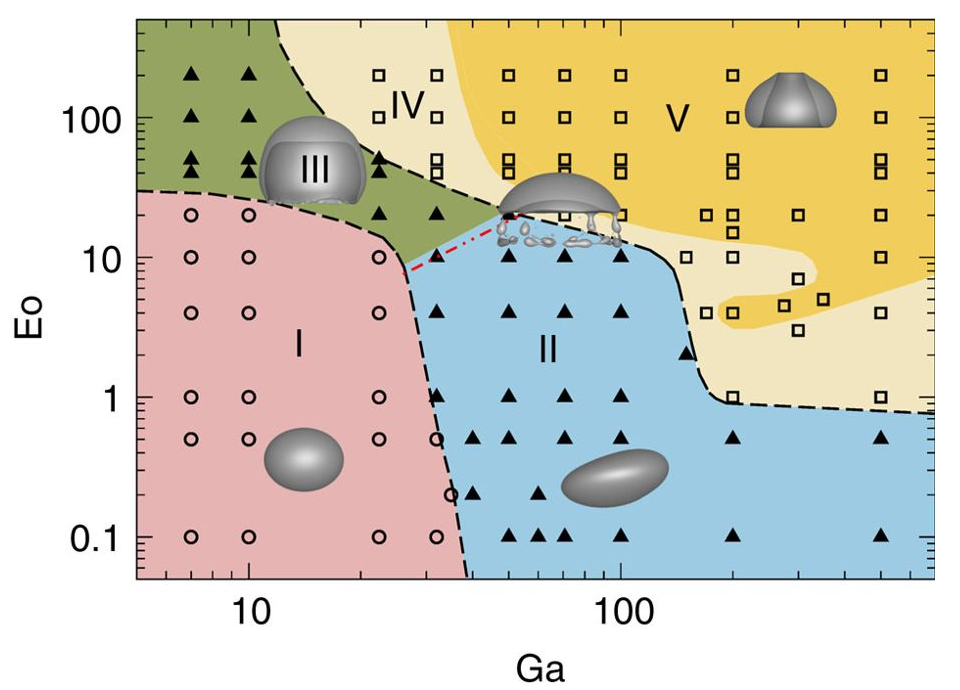
Source: M. K. Tripathi et al. 2015, figure 1.
Potential issues:
- Criterion for the boundaries?
- How to deploy?
- Higher dimensions?
- ...
Potential solution:
supervised learning
Forces acting on a rising bubble
Why does a bubble rise?
- surface tension forces
- viscous forces
- buoyancy forces
- inertia forces
- more than one force
What limits the rise velocity?
- surface tension forces
- viscous forces
- buoyancy forces
- inertia forces
- more than one force
Why are "small" bubbles spherical/ellipsoidal?
- surface tension forces
- viscous forces
- buoyancy forces
- inertia forces
- more than one force
Eötvös or Bond number
$$ Eo = \frac{\text{buoyancy force}}{\text{surface tension force}} = \frac{g\rho_l d_b^2}{\sigma} $$
$\rho_l$ - density liquid, $g$ - gravity, $d_b$ - diameter, $\sigma$ - surface tension
Tells us how strong the deformation is going to be.
Galilei number
$$ Ga = \frac{\text{inertia force}}{\text{viscous force}}\times\sqrt{\frac{\text{buoyancy force}}{\text{inertia force}}} = \frac{\sqrt{gd_b^3}}{\nu_l} $$
$\nu_l$ - kinematic liquid viscosity
Tells us how stable/unstable the rise is going to be.

Source: M. K. Tripathi et al. 2015, figure 1.
Loading, inspecting, and preparing the data

Source: M. K. Tripathi et al. 2015, figure 1.
Data extraction with Engauge Digitizer
- take screenshot of graph/figure
- load screenshot in digitizer
- set origin, axes, and scale
- select markers or lines
- extract to (csv) file
Publish your data (if you can)!
Loading and processing the data.
data_path = "../datasets/path_shape_regimes/"
regimes = ["I", "II", "III", "IV", "V"]
raw_data_files = [f"regime_{regime}.csv" for regime in regimes]
files = [pd.read_csv(data_path + file_name, header=0, names=["Ga", "Eo"]) for file_name in raw_data_files]
for data, regime in zip(files, regimes):
data["regime"] = regime
data = pd.concat(files, ignore_index=True)
data.sample(5)
# output
Ga Eo regime
22.6060 40.557 III
50.0910 39.896 V
6.9836 51.882 III
518.2900 19.694 V
101.0630 19.373 IV
Raw extracted data.
$Ga$ and $Eo$ histograms of raw data.
Data transformed using logarithm and $[-1, 1]$ scaling.
$\tilde{Ga}$ and $\tilde{Eo}$ histograms of transformed data.
Adding an ordinal label representation.
# creating an ordinal representation
logData["ordinal"] = 0.0
for i, r in enumerate(regimes):
logData.ordinal.mask(logData.regime == r, float(i), inplace=True)
Binary classification
Goal: distinguish between region I and II.
Only regions I and II
$ \tilde{Ga} = \mathrm{scale}(log(Ga)) $, $ \tilde{Eo} = \mathrm{scale}(log(Eo)) $
$$ z(\tilde{Ga}, \tilde{Eo}) = w_1\tilde{Ga} + w_2\tilde{Eo} + b $$
$$ H(z (\tilde{Ga}, \tilde{Eo})) = \left\{\begin{array}{lr} 0, & \text{if } z \leq 0\\ 1, & \text{if } z \gt 0 \end{array}\right. $$
Variation of weight for $\tilde{Ga}$.
Variation of weight for $\tilde{Eo}$.
Variation of weight for bias $b$.
Heaviside function applied to $z(\tilde{Ga} , \tilde{Eo})$.
Compact notation
Linearly weighted inputs $$ z_i=z(\mathrm{x}_i)=\sum\limits_{j=1}^{N_f}w_jx_{ij} $$
with $$ \mathbf{x}_i = \left[ \tilde{Ga}_i, \tilde{Eo}_i, 1 \right],\quad \mathbf{w} = \left[ w_1, w_2, b \right]^T $$
Binary encoding
True label: $$ y_i = \left\{\begin{array}{lr} 0, & \text{for region I }\\ 1, & \text{for region II} \end{array}\right. $$
Predicted label: $$ \hat{y}_i = H(z_i) = \left\{\begin{array}{lr} 0, & \text{if } z_i \leq 0\\ 1, & \text{if } z_i \gt 0 \end{array}\right. $$
Loss function
$$ L(\mathbf{w}) = \frac{1}{2}\sum\limits_{i=1}^N \left(y_i - \hat{y}_i\right)^2 $$The term in parenthesis can take on the values
$1$, $0$, or $-1$.
Gradient decent
Simple update rule for the weights $$ \mathbf{w}^{n+1} = \mathbf{w}^n - l_r \frac{\partial L(\mathbf{w})}{\partial \mathbf{w}} = \begin{pmatrix}w_1^n\\ w_2^n\\ b^n \end{pmatrix} + l_r \sum\limits_{i=1}^N \left(y_i - \hat{y}_i^n \right) \begin{pmatrix}\tilde{Ga}_i\\ \tilde{Eo}_i\\ 1 \end{pmatrix} $$
$n$ - current iteration, $l_r$ - learning rate
Implementation of the perceptron algorithm.
class Perceptron(object):
def __init__(self, n_weights: int):
self._p = pt.rand(n_weights)*2.0 - 1.0
def _loss(self, X: pt.Tensor, y: pt.Tensor) -> pt.Tensor:
return 0.5 * pt.sum((y - self.predict(X))**2)
def _loss_gradient(self, X: pt.Tensor, y: pt.Tensor) -> pt.Tensor:
delta = y - self.predict(X)
return -pt.cat((X, pt.ones(X.shape[0]).unsqueeze(-1)), dim=-1).T.mv(delta)
def train(self, X: pt.Tensor, y: pt.Tensor, epochs: int=500,
lr: float=0.01, tol: float=1.0e-6) -> List[float]:
loss = []
for e in range(epochs):
self._p -= lr*self._loss_gradient(X, y)
loss.append(self._loss(X, y).item())
if loss[-1] < tol:
print(f"Converged after {e+1} epochs.")
return loss
print(f"Training did not converge within {epochs} epochs")
print(f"Final loss: {loss[-1]:2.3f}")
return loss
def predict(self, X: pt.Tensor):
weighted_sum = X.mv(self._p[:-1]) + self._p[-1]
return pt.heaviside(weighted_sum, pt.tensor(0.0))
Perceptron learning rule - loss over epochs.
Perceptron algorithm - weighted sum of inputs and prediction.
Guaranteed convergence (zero loss)?
- yes
- no
- data dependent
Conditional probabilities
$$ p(y=1 | \mathrm{x}) $$speak: probability $p$ of the event $y=1$ given $\mathrm{x}$
$$ \hat{y} = f_{\mathbf{w}}(\mathrm{x}) = p(y=1 | \mathrm{x})$$model predicts class probability instead of class!
What is the expected value of $p(y=1 | \mathrm{x})$ for points far in region II?
- close to zero
- about 0.5
- close to one
What is the expected value of $p(y=1 | \mathrm{x})$ for points far in region I?
- close to zero
- about 0.5
- close to one
What is the expected value of $p(y=1 | \mathrm{x})$ for points close to the decision boundary?
- close to zero
- about 0.5
- close to one
What is the expected value of $p(y=0 | \mathrm{x})$ for points far in region I?
- close to zero
- about 0.5
- close to one
How to convert $z(\mathrm{x})$ to a probability?
- sinus: $\mathrm{sin}(z)$
- hyperbolic tangents: $\mathrm{tanh}(z)$
- sigmoid: $\sigma(z)$
Sigmoid function $\sigma = 1 / (1+\mathrm{exp}(-z))$.
Probabilities $p(II|\tilde{Ga},\tilde{Eo}) = \sigma (z(\tilde{Ga},\tilde{Eo}))$.
Joint probabilities - likelihood function
$$ l(\mathbf{w}) = \prod\limits_i^{N_s} p_i(y_i | \mathbf{x}_i) $$principle of maximum likelihood
$$ \mathbf{w}^* = \underset{\mathbf{w}}{\mathrm{argmax}}\ l(\mathbf{w}). $$Which of the following is equivalent to $l(\mathbf{w}) = \prod\limits_i^{N_s} p_i(y_i | \mathbf{x}_i)$?
- $$ \prod\limits_i^{N_s} \left[ y_i^{\hat{y}_i} (1-y_i)^{(1-\hat{y}_i)}\right] $$
- $$ \prod\limits_i^{N_s} \left[ \hat{y}_i^{y_i} (1-\hat{y}_i)^{(1-y_i)}\right] $$
Log-likelihood and binary cross entropy
$$ \mathrm{ln}(l(\mathbf{w})) = \sum\limits_{i=1}^{N_s} y_i \mathrm{ln}(\hat{y}_i) + (1-y_i) \mathrm{ln}(1-\hat{y}_i) $$
$$ L(\mathbf{w}) = -\frac{1}{N_s}\sum\limits_{i=1}^{N_s} y_i \mathrm{ln}(\hat{y}_i) + (1-y_i) \mathrm{ln}(1-\hat{y}_i) $$
Implementation of logistic regression.
class LogisticRegression(object):
def __init__(self, n_weights):
self._p = pt.rand(n_weights)*2.0 - 1.0
def _joint_probability(self, X: pt.Tensor, y: pt.Tensor) -> pt.Tensor:
probs = self.probability(X)
joint = pt.pow(probs, y) * pt.pow(1.0-probs, 1-y)
return pt.prod(joint)
def _loss(self, X: pt.Tensor, y: pt.Tensor) -> pt.Tensor:
probs = self.probability(X)
entropy = y*pt.log(probs+1.0e-6) + (1-y)*pt.log(1.0-probs+1.0e-6)
return -entropy.mean()
def _loss_gradient(self, X: pt.Tensor, y: pt.Tensor) -> pt.Tensor:
delta = y - self.probability(X)
grad = pt.cat((X, pt.ones(X.shape[0]).unsqueeze(-1)), dim=-1) * delta.unsqueeze(-1)
return -grad.mean(dim=0)
def train(self, X: pt.Tensor, y: pt.Tensor, epochs: int=10000,
# snip - see perceptron
def probability(self, X: pt.Tensor) -> pt.Tensor:
weighted_sum = X.mv(self._p[:-1]) + self._p[-1]
return pt.sigmoid(weighted_sum)
def predict(self, X: pt.Tensor) -> pt.Tensor:
return pt.heaviside(self.probability(X)-0.5, pt.tensor(0.0))
Binary cross entropy vs. epochs.
Landscape around the optima of binary cross entropy and joint probability.
Probability and prediction with logistic regression.
New goal: separate regime I from II and III.
Logistic regression classifiers separating regions I-II and I-III.
Training of two logistic regressors.
combined_prob = pt.sigmoid(
0.9*model_I_II.probability(pt.stack((xx.flatten(), yy.flatten())).T)
+ 0.9*model_I_III.probability(pt.stack((xx.flatten(), yy.flatten())).T)
- 0.5
)
combined_prediction = pt.heaviside(combined_prob - 0.5, pt.tensor(0.0))
Combination of two logistic regression classifiers.
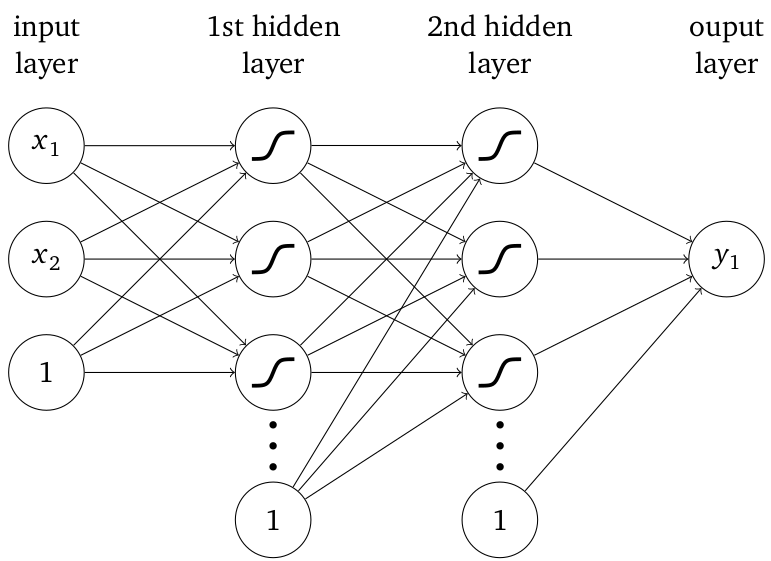
Generic neural network with fully connected layers.
Multi-class classification
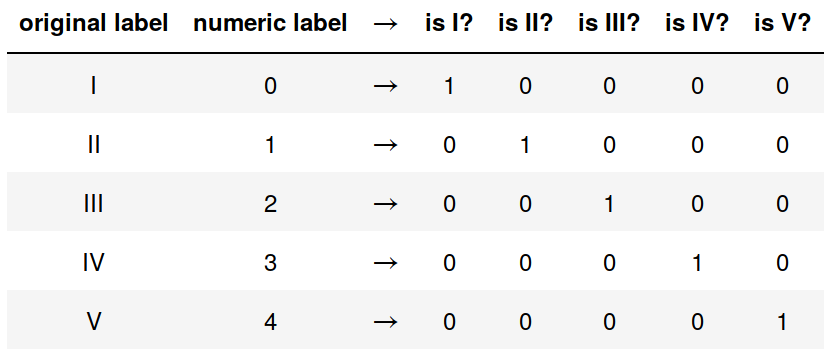
One hot encoding of path regimes.
What is the length of the label vector if we have 10 classes (regimes)?
- 1
- 2
- 5
- 10
What does the label vector look like if we have 4 classes and the true label is class 3 / regime 3 (start counting from 1)?
- $\mathbf{y}_i = \left[ 1, 0, 0, 0 \right]^T$
- $\mathbf{y}_i = \left[ 0, 0, 1, 0 \right]^T$
- $\mathbf{y}_i = \left[ 1, 0, 1, 0 \right]^T$
- $\mathbf{y}_i = \left[ 0, 0, 3, 0 \right]^T$
Softmax function and categorical cross entropy for $K$ classes:
$$ p(y_{j}=1 | \mathbf{x}) = \frac{e^{z_{j}}}{\sum_{j=0}^{K-1} e^{z_{j}}} $$
$$ L(\mathbf{w}) = -\frac{1}{N} \sum\limits_{i=1}^{N_s}\sum\limits_{j=0}^{K-1} y_{ij} \mathrm{ln}\left( \hat{y}_{ij} \right) $$
What is a likely/possible prediction if the true class is 3 (start counting from 1)?
- $\hat{\mathbf{y}}_i = \left[ 0.1, 0, 0.9, 0.1 \right]^T$
- $\hat{\mathbf{y}}_i = \left[ 1, 0, 0, 0 \right]^T$
- $\hat{\mathbf{y}}_i = \left[ 0, 0.02, 0.95, 0.03 \right]^T$
- $\hat{\mathbf{y}}_i = \left[ 0, 0, 3, 0 \right]^T$
Neural network classification model.
n_features, n_classes, n_neurons = 2, 5, 50
regime_model = pt.nn.Sequential(
pt.nn.Linear(n_features, n_neurons),
pt.nn.Tanh(),
pt.nn.Linear(n_neurons, n_neurons),
pt.nn.Tanh(),
pt.nn.Linear(n_neurons, n_classes)
)
Training loop definition.
dataset = pt.utils.data.TensorDataset(
pt.tensor(logData[["Ga", "Eo"]].values, dtype=pt.float32),
pt.tensor(logData.ordinal.values, dtype=pt.int64)
)
train_size = int(0.9*len(dataset))
val_size = len(dataset) - train_size
train_dataset, val_dataset = pt.utils.data.random_split(dataset, (train_size, val_size))
train_loader = pt.utils.data.DataLoader(train_dataset, batch_size=len(train_dataset), shuffle=True)
val_loader = pt.utils.data.DataLoader(val_dataset, batch_size=len(val_dataset))
results = train_model(regime_model, pt.nn.CrossEntropyLoss(), train_loader, val_loader,
epochs=1000, optimizer=optimizer)
Categorical cross entropy vs. epochs.
Prediction of classification network.
